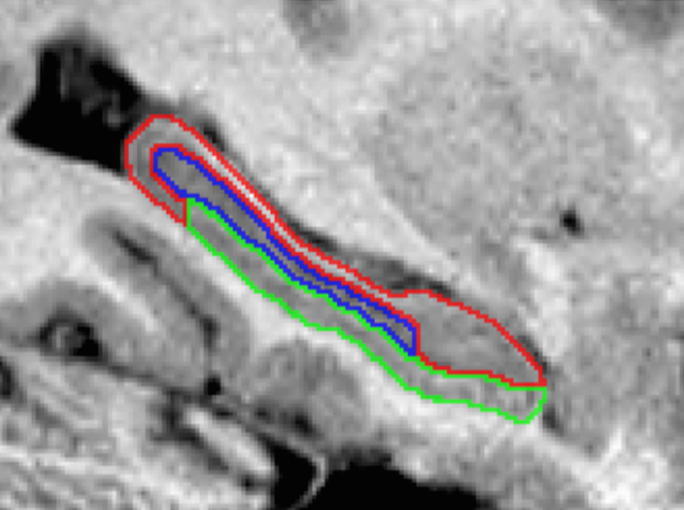
Summary: The hippocampus is composed of distinct anatomical subregions that participate in multiple cognitive processes and are differentially affected in prevalent neurological and psychiatric conditions. Advances in high-field MRI allow for the non-invasive identification of hippocampal substructure. These approaches, however, demand time-consuming manual segmentation that relies heavily on anatomical expertise. Here, we share manual labels and associated high-resolution MRI data (MNI-HISUB25; submillimetric T1- and T2-weighted images, detailed sequence information, and stereotaxic probabilistic anatomical maps) based on 25 healthy subjects. Data were acquired on a widely available 3-Tesla MRI system using a 32 phased-array head coil. The protocol divided the hippocampal formation into three subregions: subicular complex, Cornu Ammonis 1, 2 and 3 (CA1-3), and CA4-dentate gyrus (CA4-DG). Segmentation was guided by consistent intensity and morphology characteristics of the densely myelinated molecular layer together with few geometry-based boundaries flexible to overall mesiotemporal anatomy, and achieved excellent intra-/inter-rater reliability (Dice index ≥90/87%). The dataset can inform neuroimaging assessments of the mesiotemporal lobe and help to develop segmentation algorithms relevant for basic and clinical neurosciences.
MRI data were acquired on a 3-Tesla Siemens TimTrio scanner using a 32-channel head coil. We obtained two sets of T1w images: a 3D magnetization-prepared rapid-acquisition gradient echo (MPRAGE) with millimetric resolution (repetition time (TR)=2300 ms; echo time (TE)=2.98 ms; inversion time (TI)=900 ms; flip angle=9°; matrix size=256×256; field-of-view (FOV)=256×256 mm2; 176 sagittal slices with 1 mm slice thickness resulting in 1×1×1 mm3 voxels; iPAT=2, acquisition time=5.30 min), and a submillimetric 3D MPRAGE (TR=3000 ms; TE=4.32 ms; TI=1500 ms; flip angle=7°; matrix size=336×384; FOV=201×229 mm2; 240 axial slices with 0.6 mm slice thickness resulting in 0.6×0.6×0.6 mm3 voxels; acquisition time=16.48 min; to increase the signal-to-noise ratio, two identical scans were acquired, motion corrected, and averaged into a single volume). T2w images were obtained using a 2D turbo spin-echo sequence (TR=10810 ms; TE=81 ms; flip angle=119°; matrix size=512×512; FOV=203×203 mm2, 60 coronal slices angled perpendicular to the hippocampal long axis, slice thickness=2mm, resulting in 0.4×0.4×2.0 mm3 voxels; acquisition time=5.47 min).
This dataset includes:
Authors:
USAGE AGREEMENT
Creative Commons License: Attribution - Non-Commercial
Funding
This work and BCB were supported by the Canadian Institutes of Health Research (CIHR MOP-57840 and CIHR MOP-123520).
| Imaging_Data |
NITRC Download Instructions
In order to access the data, you must first register for an account with NITRC. After you have done so, click here to request access to the 1000 Functional Connectomes Project on NITRC. Users will be approved within 1 business day.
Publications:
J. Kulaga-Yoskovitz, B.C. Bernhardt, S. Hong, T. Mansi, K. Liang, A. van der Kouve, J. Smallwood, A. Bernasconi, N. Bernasconi (2015) Multi-contrast and submillimetric 3T hippocampal subfield segmentation protocol and dataset. Scientific data, in press.